|
- Home
- BioZoom
- BioZoom archives
- 2018
- 2018 Issue 4
- How CRISPR Changed Drug Development
How CRISPR Changed Drug Development |
Author: Lasse Ebdrup Pedersen, Senior Researcher, DTU Biosustain, Novo Nordisk Foundation Center for Biosustainability, laeb@biosustain.dtu.dk |
Chinese Ovary Hamster cells are widely used for the bioproduction of therapeutic recombinant proteins. However, the creation and optimization of new bioproduction cell lines is laborious and expensive. CRISPR is a revolutionizing tool that allows for specific, targeted changes to the cell factories that can substantially increase the effectiveness of protein production. The best selling drugs (money per year) in the world are primarily recombinant therapeutic proteins. These drugs are used to treat a variety of diseases such as rheumatoid arthritis, Crohn’s disease, and breast cancer. The key reason these drugs are the highest selling is not due to extreme demand, but rather the high cost per treated patient. The high cost is putting health care systems under pressure because an increasingly larger percentage of their budgets now have to be spent on medicine itself. Drug production in Chinese Hamster Ovary cells The bioproduction of recombinant therapeutic proteins is most commonly done in large cell cultures and the cell host of choice is the Chinese Hamster Ovary (CHO) cell. Originally, this choice of host cell was among others due to its ability to produce large complex proteins with mammalian posttranslational modifications. Ever since, the CHO cell has been building up a decades long history of safety and regulatory approval as well as been the platform for development of molecular biology tools and bioprocess expertise. Bioproduction of large quantities of a therapeutic protein in CHO cells is a workhorse in the pharmaceutical indstry. However, the process of making proteins with the correct quality attributes has largely been treated as a black box engineering problem: 1) Insert the gene of interest (GOI) encoding the therapeutic protein into one or more random positions in the genome. 2) Sometimes random mutagenesis will be employed to create more diversity. 3) Then screen thousands of cells for desirable properties both regarding the therapeutic protein as well as the cell line itself, and 4) then develop an optimal bioprocess. This process leaves a very large degree of uncertainty regarding how long, if at all possible, it will take to go from inserting a gene of interest (GOI ) to actually having an economically viable, regulatory affairs approvable, protein production going. This is part of the reason for the high price of recombinant therapeutic proteins. Rational improvement of gene engineering approaches The CHO program here at the Novo Nordisk Foundation Center for Biosustainability (CFB) at DTU has as its central mission to improve our understanding of the CHO cell to enable a rational engineering approach. Three groups are involved in the research:a scientific group, a translational core group, and a computational group. We started our work almost 6 years ago and have focused on four main projects:
Taken together this work would not have been possible without CRISPR/Cas9 gene editing. How CRISPR revolutionized gene engineering of CHO cell lines The CRISPR/Cas9 genome editing technology was a perfectly timed breakthrough for our needs. In the autumn of 2012 we were looking at the various technologies available for genome editing. We, together with several other groups, had just sequenced the genome of the CHO cell line ‘K1’(1) which would enable us to use the genomic editing tools: TALENS and Zinc fingers (Znf). However we found the frequency of succesful editing was very low, usually less than 1% of cells had the desired edits. Moreover, it was very expensive, several thousand dollars for 1 Znf pair, which equals e.g. one gene knockout (KO). Luckily, our center also hosts several groups working with bacteria as cell factories and, over beers, a talented PhD student, Carlotta Ronda, told me about a recent talk she attended describing a novel RNA programmable DNA nuclease (Cas9) in adaptive bacterial immunity (the CRISPR/Cas9 system). This method demonstrated that using two pieces of RNA (later one, a gRNA), one could guide the Cas9 nuclease to a specific location on the genome where it would then perform a double stranded cut. After such a cut, a mammalian cell will attempt to repair it, which occasionally results in the generation of an indel, a few basepairs added or removed at the location of the break. This can result in a frameshift which renders the gene KO. We quickly reasoned that it was worth testing whether this system could be used in mammalian cells like CHO cells. It could. Others beat us to the publication and fame, but like everybody else, we found that CRISPR/Cas9 had excellent editing efficiency and equally important, was extremely cheap to use. We then established methods to use multiple gRNAs simultaneously and gene knock-ins (6) and we are currently working on genome wide libraries of gRNAs that allow us to efficiently elucidate pathways and screen for genes involved in phenotypes of interest. Using our genome editing methods we have created over 500 CHO cell lines with a variety of phenotypes of interest. CRISPR/Cas9 as a tool to optimize specific production parameters We had a hypothesis that we could make a CHO cell, which produces more of our recombinant therapeutic if we removed unnecessary genes that occupied the translational machinery. To test this theory we obtained a CHO cell producing a therapeutic antibody and subjected it to ribosome profiling to identify what occupied the ribosomes (Figure 1). We could see that the antibody heavy and light chains did indeed occupy a large fraction of ribosomes, but to our surprise, an antibiotic selection marker, NeoR, that had been used to establish the cell line, was the second highest occupant. Knocking down the NeoR gene resulted in improved protein production (9). 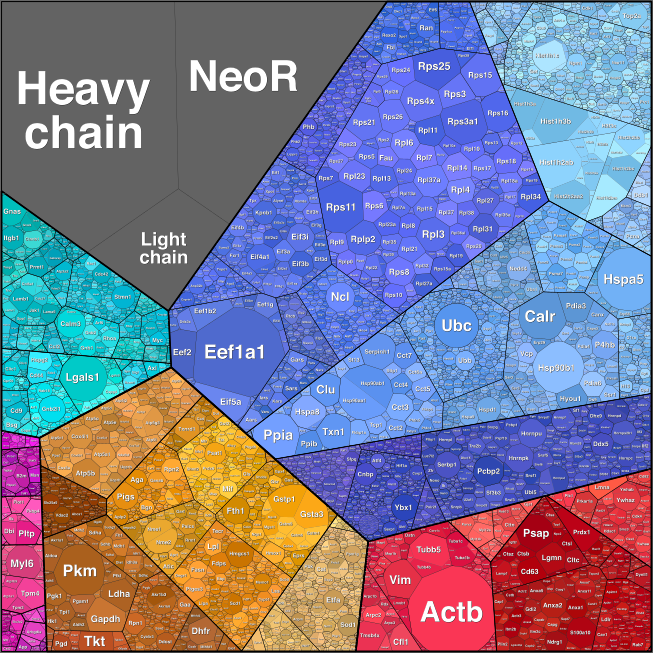
Figure 1: Ribosomal occupancy in a CHO cell producing an antibody (heavy and light chain) and an antibiotic selection marker (NeoR). Having demonstrated that the general hypethesis held true, we extended the purpose of the project to clean up the secretory pathway for the dual purpose of freeing up translational capacity as well as lower the amount of work required to purify the secreted therapeutic protein. We identified what the CHO cell was secreting using mass spectrometry and then we used Cas9 to knock out the most common pollutants. This is an ongoing project where we continually knock out more and more genes (unpublished, patent pending). In another project we used CRISPR/Cas9 to modify the glycosylation of secreted proteins. Protein glycosylation plays important roles in solubility, stability and function of secreted proteins. Non-human glycosylation can also provoke an immune reaction potentially leaving the patient allergic to life saving medicine. For these reasons, controlling protein glycosylation is of upmost importance. An example of our work in this area is shown in recently published study where we demonstrated multi KO cell lines that only have ~1% of its total secreted proteins that are N-galactosylated (7). These cell lines can replace expensive post-production enzymatic steps to achieve the same glycoform (10), potentially lowering production costs. References 1. X. Xu et al., “The genomic sequence of the Chinese hamster ovary (CHO)-K1 cell line.,” Nat. Biotechnol., vol. 29, no. 8, pp. 735–41, Jul. 2011. 2. A. Singh, H. F. Kildegaard, and M. R. Andersen, “An Online Compendium of CHO RNA-Seq Data Allows Identification of CHO Cell Line-Specific Transcriptomic Signatures,” Biotechnol. J., p. 1800070, Jul. 2018.3. H. Hefzi et al., “A Consensus Genome-scale Reconstruction of Chinese Hamster Ovary Cell Metabolism,” Cell Syst., vol. 3, no. 5, p. 434–443.e8, Nov. 2016. 4. J. M. Gutierrez et al., “Genome-scale reconstructions of the mammalian secretory pathway predict metabolic costs and limitations of protein secretion,” bioRxiv, p. 351387, Jun. 2018. 5. P. N. Spahn, A. H. Hansen, H. G. Hansen, J. Arnsdorf, H. F. Kildegaard, and N. E. Lewis, “A Markov chain model for N-linked protein glycosylation – towards a low-parameter tool for model-driven glycoengineering,” Metab. Eng., vol. 33, pp. 52–66, Jan. 2016. 6. J. S. Lee, T. B. Kallehauge, L. E. Pedersen, and H. F. Kildegaard, “Site-specific integration in CHO cells mediated by CRISPR/Cas9 and homology-directed DNA repair pathway,” Sci. Rep., vol. 5, no. 1, pp. 1–11, Jul. 2015. 7. T. Amann, A. H. Hansen, S. Kol, G. M. Lee, M. R. Andersen, and H. F. Kildegaard, “CRISPR/Cas9-Multiplexed Editing of Chinese Hamster Ovary B4Gal-T1, 2, 3, and 4 Tailors N -Glycan Profiles of Therapeutics and Secreted Host Cell Proteins,” Biotechnol. J., Jul. 2018. 8. L. M. Grav et al., “Minimizing Clonal Variation during Mammalian Cell Line Engineering for Improved Systems Biology Data Generation,” ACS Synth. Biol., p. acssynbio.8b00140, Aug. 2018. 9. T. B. Kallehauge et al., “Ribosome profiling-guided depletion of an mRNA increases cell growth rate and protein secretion.,” Sci. Rep., vol. 7, no. 1, p. 40388, Dec. 2017. 10. G. Dekkers et al., “Multi-level glyco-engineering techniques to generate IgG with defined Fc-glycans,” Sci. Rep., vol. 6, no. 1, p. 36964, Dec. 2016. |
Looking for something? Search the website here: |
